Wei YuYuming MaSharmila ShankarRakesh K. Srivastava
Abstract
The incidence of pancreatic cancer is on the rise. Risk factors for pancreatic cancer include alcohol toxicity and metabolic conditions such as obesity, hypertension, dyslipidaemia, insulin resistance and type 2 diabetes. However, the molecular mechanism by which chronic alcohol consumption contributes to pancreatic cancer is not well understood. The purpose of the study was to demonstrate the effects of long‐term chronic ethanol exposure on the transformation of human pancreatic normal ductal epithelial (HPNE ) cells. Our data showed that ethanol‐transformed HPNE cells were more progressively transformed exhibiting spheroids and colonies, and anchorage‐independent growth. These transformed cells contained high levels of reactive oxygen species and induced SATB 2 expression. Furthermore, during ethanol‐induced cellular transformation, cells gained the phenotypes of cancer stem cells (CSC s) by expressing pluripotency maintaining factors (Oct4, Sox2, cM yc and KLF 4) and stem cell markers (CD 24, CD 44 and CD 133). Ethanol‐induced SATB 2 can bind to the promoters of KLF 4, Oct4, cM yc, Sox2, Bcl‐2 and XIAP genes. Suppression of SATB 2 expression in ethanol‐transformed HPNE cells inhibited cell proliferation, colony formation and markers of CSC s and pluripotency. These data suggest that chronic alcohol consumption may contribute toward the development of pancreatic cancer by converting HPNE cells to cancer stem‐like cells.
1 INTRODUCTION
Pancreatic cancer has an exceptionally high mortality rate and is the fourth leading cause of cancer‐related death in the United States.1 With an overall 5‐year survival rate of 6%,2 pancreatic cancer has one of the poorest prognoses among all cancers.3 The incidence of pancreatic cancer varies significantly across regions, which suggests that several factors may be responsible for this deadly disease.4 The genetic, race, gender, environmental carcinogen, diet and lifestyle appears to be the primary factors for pancreatic cancer.5 Other factors such as smoking, alcohol, coffee consumption and exposure to organochlorine or hydrocarbon solvent have been associated with the frequency and spectrum of K‐ras mutation in pancreatic tumours.6–9 Metabolic conditions such as obesity, hypertension, dyslipidaemia, insulin resistance, and type 2 diabetes are also the risk factors for pancreatic cancer.8, 10 About 5%‐10% of patients with pancreatic cancer have underlying germline mutations or disorders, whereas the remaining percentage of cancer cases may be because of somatic mutations.4
Epidemiological data strongly suggest that the heavy alcohol drinking increases the risk for pancreatic cancer.11–14 A recent study has demonstrated that chronic alcohol intake promotes intestinal tumorigenesis and tumour invasion in genetically susceptible mice, increases in polyp‐associated mast cells, and mast‐cell‐mediated tumour migration in vitro,15 suggesting mast‐cell‐mediated inflammation could promote carcinogenesis.15 These data confirm that ethanol and its metabolites are the potential human carcinogen. However, the molecular mechanism by which ethanol toxicity induces malignant transformation of human pancreatic normal ductal epithelial (HPNE) cells is not known.
SATB2 (special AT‐rich binding protein‐2), a transcription factor and epigenetic regulator that binds DNA 16 to regulate gene expression.17–19 SATB2 gene is required for normal mammalian development; however, it is not expressed in healthy adult cells. SATB2 is essential for proper facial patterning of the embryo and healthy bone development.19 Inappropriate activation of this gene may be the cause of malignant cellular transformation. SATB2 regulates transcription of pluripotency maintaining factors (Sox2, cMyc, KLF4 and Oct4) which form the core regulatory positive feedback‐loop to sustain self‐renewal capacity of stem cells. Using chromatin immunoprecipitation assay, we have shown that SATB2 can directly bind to the promoters of Bcl‐2, Bsp, Nanog, cMyc, XIAP, KLF4 and Hoxa2, suggesting a role of SATB2 in the regulation of cell survival, pluripotency and proliferation.20 Therefore, we reasoned to believe that SATB2 proteins may play a critical role during chronic ethanol exposure of human pancreatic ductal epithelial cells.
The primary goal of this paper was to examine the molecular mechanisms by which chronic ethanol exposure induces cellular transformation of HPNE cells which may lead to pancreatic carcinogenesis. To investigate the role of SATB2 at an early step of cell transformation, we utilized HPNE cells as a model to generate progenitor cells by chronic ethanol exposure. Our studies have established a novel link between exposure to ethanol and SATB2‐regulated transformation of HPNE cells.
2 MATERIALS AND METHODS
2.1 Cell culture conditions and reagents
Human pancreatic normal ductal epithelial cells were purchased from American Type Culture Collection, Manassas, VA. HPNE cells were grown in well‐defined cell medium as described.21 Antibodies against SATB2 and β‐actin were purchased from Abcam (Cambridge, MA). Enhanced chemiluminescence (ECL) Western blot detection reagents were purchased from Amersham Life Sciences Inc. (Arlington Heights, IL).
2.2 Cell proliferation assay
Cells (1.5 × 104) were incubated for various time points in 1 mL of culture medium. Cell viability was determined by trypan blue assay using Countess™ Automated Cell Counter (Invitrogen).
2.3 Colony formation assay
Colony formation assays were performed as described elsewhere.22 In brief, cells were seeded into 6‐well plates at a low density (200 cells per well). Cell culture medium was renewed every 3 days. After 21 days, colonies were fixed with cold methanol and then stained with 0.5% crystal violet. The colonies were imaged with a microscope.
2.4 Spheroid formation
Spheroids formation assays were performed as described elsewhere.22 In brief, cells were plated in ultra‐low attachment plates at a density of 100‐500 cells/mL. The spheroid formation in suspension was measured after 10 days of culture using a Nikon Eclipse microscope (Nikon).
2.5 Lentiviral particle production and transduction
The lentivirus production and transduction were performed as described elsewhere.22 In brief, lentivirus was produced by triple transfection of HEK 293T cells. Packaging 293T cells were plated in 10‐cm plates at a cell density of 5 × 106 1 day before transfection in DMEM containing 10% heat‐inactivated foetal bovine serum. 293T cells were transfected with 4 μg of plasmid and 4 μg of the lentiviral vector using lipid transfection (Lipofectamine‐2000/Plus reagent; Invitrogen) according to the manufacturer’s protocol. Viral supernatants were collected and concentrated by adding PEG‐it virus precipitation solution (SBI System Biosciences) to produce virus stocks with titres of 1 × 108‐1 × 109 infectious units per mL. Viral supernatant was collected for 3 days by ultracentrifugation and concentrated 100‐fold. Titres were determined on 293T cells. Cells were transduced with lentiviral particles expressing the gene of interest.
2.6 Western blot analysis
The Western blot analysis was performed as we described earlier.23 In brief, cell lysates were subjected to SDS‐PAGE, and gels were blotted onto nitrocellulose membrane (Amersham Biosciences, Piscataway, NJ, USA). The membranes were blocked with 5% BSA in Tris‐Tween buffered saline at 37°C for 2 hours and then incubated with primary antibody diluted in Tris‐buffered saline (1:1000 dilutions) overnight at 4°C, with gentle shaking. The membranes were then washed 3 times with Tris‐buffered saline‐T (TBS‐T) and incubated with the secondary antibody linked to horseradish peroxidase (1:5000) for 1 hour. After incubation with secondary antibody, the membranes were rewashed 3 times with TBS‐T. Finally, protein‐antibody complexes were detected by the addition of ECL substrate (Thermo Fisher Scientific, Rockford, IL).
2.7 Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were performed as we described elsewhere.24, 25 In brief, chromatin was immunoprecipitated using anti‐SATB2 antibody. Normal rabbit IgG (Abcam) was used as a negative control. ChIP‐derived DNA was measured by 2% agarose gel electrophoresis.
- Bcl‐2 promoterChIP‐F, TTTCAGCATCACAGAGGAAG
- Bcl‐2 promoterChIP‐R, CAATCACGCGGAACACTTGATT
- Oct4‐ChIP‐F, ATGACCACTGCGCCCGGACTGC
- Oct4‐ChIP‐R, ACTTGGATCTCTTCCAAGTGC
- KLF4‐Chip‐F, ACCGGACCTACTTACTCGCC
- KLF4‐Chip‐R, TCGGCAGCCCGAAGCAGCTGG
- cMyc‐Chip‐F, AATTAATGCCTGGAAGGCAGCC
- cMyc‐ChIP‐R, AGTCAGCAGAGACCCTTGTG
- XIAP‐Chip‐F,TCCAAGAGAGATGCACTAGGGTC
- XIAP‐Chip‐R, TTATGGCAAGATCTATGTGGAACTC
2.8 Quantitative real‐time PCR
Total RNA in cells was extracted by the TRIzol reagent (Invitrogen) and reverse transcribed into cDNA using High‐Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). qRT‐PCR was conducted using fast SYBR Green Master Mix (Thermo Fisher Scientific). The 2−ΔΔCt method was used to evaluate relative mRNA expressions compared with controls. The following gene‐specific primers were used:
- Sox2 (5′‐AAC CCC AAG ATG CAC AAC TC‐3′, 5′‐GCT TAG CCT CGT CGA TGA AC‐3′)
- cMyc (5′‐CGA CGA GAC CTT CAT CAA AA‐3′, 5′‐TGC TGT CGT TGA GAG GGT AG‐3′)
- Oct4 (5′‐GGA CCA GTG TCC TTT CCT CT‐3′, 5′‐CCA GGT TTT CTT TCC CTA GC‐3′)
- CD24 (5′‐ATG GGA ACA AAC AGA TCG AA‐3′, 5′‐TTT GCT CTT TCA GCC ATT TC‐3′)
- CD44 (5′‐ACT TCA CCC CAC AAT CTT GA‐3′, 5′‐GTG GCT TGT TGC TTT TCA GT‐3′)
- CD133 (5′‐CCT CTG GTG GGG TAT TTC TT‐3′, 5′‐CCT CTG GTG GGG TAT TTC TT‐3′)
- HK‐GAPD (5′‐GAG TCA ACG GAT TTG GTC GT‐3′, 5′‐TTG ATT TTG GAG GGA TCT CG‐3′)
2.9 Statistical analysis
The mean and SD were calculated for each experimental group with replicates. Differences between groups were analysed by ANOVA, followed by Bonferroni’s multiple comparison tests using PRISM statistical analysis software (GrafPad Software, Inc., San Diego, CA). Significant differences among groups were calculated at P < .05.
3 RESULTS
3.1 Ethanol induces transformation of HPNE cells by up‐regulating SATB2 expression
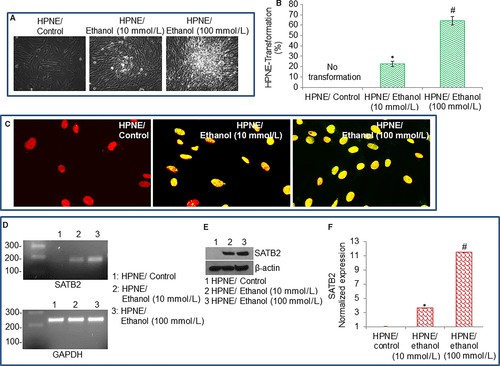
We have used HPNE cells as a model to assess whether chronic ethanol exposure induces malignant transformation. HPNE cells were grown in culture medium in the presence or absence of ethanol (10 and 100 mmol/L) for 6 months. Long‐term chronic exposure of HPNE cells to ethanol‐induced cellular transformation as evident by the formation of clumps, loss of contact inhibition, and disoriented growth (Figure 1A). HPNE cell transformation efficiency was significantly higher with the higher dose of ethanol (100 mmol/L) compared to 10 mmol/L ethanol exposure (Figure 1B).

SATB2 plays a vital role in the chromatin remodelling and regulation of genes which participates in cell growth, survival, differentiation, self‐renewal and pluripotency. We, therefore, examined the mechanism of ethanol‐induced transformation of HPNE cells by comparing the expression of SATB2 in HPNE control cells and ethanol‐transformed HPNE cells (HPNE/Ethanol). As shown in Figure 1C‐E, 6‐month exposure of HPNE cells to ethanol‐induced the expression of SATB2 gene as measured by immunohistochemistry, polymerase chain reaction, Western blotting and quantitative real‐time polymerase chain reaction. SATB2 was not expressed in normal HPNE cells. By comparison, SATB2 was expressed in the nuclei of HPNE/Ethanol cells (appearance of yellow colour), but not in HPNE/Control cells. Short‐term exposure (up to 1 month) of HPNE cells to ethanol did not induce SATB2 (data not shown). Our data suggest that ethanol can induce HPNE cell transformation which is associated with the induction of SATB2.
3.2 Ethanol‐transformed HPNE cells form spheroids in suspension and colonies in soft agar, express stem cell markers and pluripotency maintaining factors, and generate reactive oxygen species
We next examined whether ethanol‐transformed HPNE cells gained the phenotypes of cancer stem cells (CSCs) and express pluripotency maintaining markers (Figure 2). The formation of spheroids in suspension is the main characteristics of CSCs. After 6 months long exposure of HPNE cells to ethanol, HPNE cells demonstrated the features of cellular transformation, that is formed spheroids in suspension and colonies in soft agar (Figure 2A). Dimensions of spheroids and colonies formed by HPNE cells were greater with exposure of 100 mmol/L ethanol compared to 10 mmol/L ethanol. We next examined whether transformed HPNE cells gained the phenotype of CSCs by measuring stem cell markers (Figure 2B) and pluripotency maintaining factors (Figure 2C). Ethanol‐transformed HPNE cells expressed significantly higher levels of stem cell markers (CD24, CD44 and CD133) and pluripotency maintaining factors (Oct4, Sox2, cMyc and Klf4). We have validated the use of these markers and pluripotency maintaining factors for pancreatic CSCs.26–28 The induction of these genes was higher with 100 mmol/L of ethanol compared to 10 mmol/L ethanol. The expression of these genes was either low or not detected in HPNE cells. Reactive oxygen species (ROS) are generated during malignant transformation and are elevated in inflammatory environment. N ‐acetylcysteine (NAC) is an aminothiol and synthetic precursor of intracellular cysteine and glutathione (GSH) and is thus considered an important antioxidant. Ethanol‐transformed HPNE cells generated ROS (Figure 2D). Furthermore, treatment of ethanol‐transformed HPNE cells with NAC inhibited intracellular ROS levels. These data suggest that ethanol can induce transformation of HPNE cells by converting them to CSC‐like phenotype.

3.3 SATB2 directly binds to promoters of Klf4, Oct4, cMyc, Sox2, Bcl‐2 and XIAP in ethanol‐transformed HPNE cells
SATB2 is a transcription factor and thus can regulate various cellular functions by directly binding to target genes.29 Some of the target genes of SATB2 regulate pluripotency, self‐renewal, and cell survival. ChIP assay is commonly used to examine the binding of a transcription factor to the promoter regions of the target genes. We, therefore, investigated the binding of SATB2 to the promoters of potential gene targets in HPNE‐transformed cells. SATB2 can directly bind to promoters of KLF4, Oct4, cMyc, Sox2, Bcl‐2 and XIAP (Figure 3). These data suggest that SATB2 can directly bind to these genes and regulate their expression in ethanol‐transformed cells.

3.4 Knockdown of SATB2 by shRNA in ethanol‐transformed cells inhibits cell proliferation and colony formation
To examine whether SATB2 in involved in in vitro cellular transformation, we knocked‐down the expression of SATB2 by shRNA in ethanol‐transformed HPNE cells which were exposed to 10 or 100 mmol/L ethanol for 6 months. Ethanol‐transformed HPNE cells were transduced with either scrambled or SATB2 shRNA lentiviral particles, and cell growth and colony formation were measured (Figure 4). Transduction of Ethanol‐transformed HPNE cells (HPNE/Ethanol [10 or 100 mmol/L]) with SATB2 shRNA viral particles inhibited the expression of SATB2 proteins and mRNA compared to that of HPNE/Ethanol (10 or 100 mmol/L)/Scrambled cells (Figure 4A and B).

We next examined the effects of inhibiting SATB2 on cell proliferation, and colony formation by ethanol‐transformed cells (Figure 4C and D). SATB2 shRNA inhibited cell proliferation, and colony formation in both ethanol‐transformed HPNE groups (HPNE/Ethanol [10 mmol/L]/SATB2 shRNA and HPNE/Ethanol [100 mmol/L]/SATB2 shRNA) compared to that of Scrambled groups (HPNE/Ethanol [10 mmol/L]/Scrambled and HPNE/Ethanol [100 mmol/L]/Scrambled). These data suggest that SATB2 can inhibit the ability of Ethanol‐transformed cells to proliferate and form colonies.
3.5 SATB2 inhibits the expression of stem cell markers in ethanol‐transformed cells
Our data demonstrate that chronic exposure of HPNE cells with ethanol induces stem cell markers CD133, CD44 and CD24. We, therefore, sought to examine whether inhibition of SATB2 by shRNA attenuates the expression of these markers in ethanol‐transformed cells (Figure 5A‐C). SATB2 shRNA inhibited the expression of CD133, CD44 and CD24 in ethanol‐transformed cells (HPNE/Ethanol [10 mmol/L]/SATB2 shRNA, and HPNE/Ethanol [100 mmol/L]/SATB2 shRNA) compared to that of scrambled control groups. These data suggest that inhibition of SATB2 can result in suppressing stem cell population in ethanol‐transformed HPNE cells (HPNE/Ethanol).

3.6 SATB2 inhibits the expression of pluripotency maintaining factors in ethanol‐transformed cells
Since ethanol‐transformed HPNE cells demonstrated stemness by overexpressing pluripotency maintaining factors, we next sought to examine whether inhibition of SATB2 will attenuate the expression of cMyc, KLF4, Sox2 and Oct4 in ethanol‐transformed cells (Figure 6A‐D). SATB2 shRNA inhibited the expression of cMyc, KLF4, Sox2 and Oct4 in ethanol‐transformed HPNE cells (HPNE/Ethanol [10 mmol/L]/SATB2 shRNA, and HPNE/Ethanol [100 mmol/L]/SATB2 shRNA). These data suggest that SATB2 can regulate pluripotency and self‐renewal by regulating the expression of cMyc, KLF4, Sox2 and Oct4 in ethanol‐transformed HPNE cells (HPNE/Ethanol).

4 DISCUSSION
The proposed studies provide, for the first time, the carcinogenic effects of alcohol on HPNE cells. We have provided ample evidence that during ethanol‐induced cellular transformation, CSCs/progenitor cells are developed which may play a significant role in pancreatic carcinogenesis. The current findings enhance our understanding of the signalling pathways which regulate cellular transformation and examine how chronic exposure of HPNE cells to ethanol induces stem cell markers, and pluripotency maintaining factors resulting in the generation of cancer stem‐like cells. The conversion of HPNE cells to cancer stem‐like cells by ethanol pointing a pathophysiological role of alcohol abuse in pancreatic carcinogenesis. The conversion of HPNE cells to stem‐like cells by ethanol will be a significant event for those individuals who are addicted to alcohol consumption.
Alcohol abuse is a risk factor for pancreatic cancer. Genetic and other environmental factors like cigarette smoke may further potentiate the adverse effects of alcohol. On the basis of our findings, we hypothesized that ethanol toxicity induces HPNE cell transformation and may induce or enhance, alone or with other factors, pancreatic carcinogenesis by modulating the expression of SATB2. We recently showed that up‐regulation of SATB2 in HPNE cells was sufficient to induce malignant transformation and those transformed cells gained the phenotypes of CSCs by expressing CSC markers and pluripotency maintaining factors.24, 30 Chronic exposure of HPNE cells to 100 mmol/L ethanol caused ~300‐fold increase in CD133 expression, whereas knockdown of SATB2 resulted in a moderate, ~2.5‐fold reduction in CD133 levels. These data suggest the possibility of other transcription factors/genes that might contribute to regulation of ethanol‐dependent transformation processes. Furthermore, SATB2‐transformed cells also showed high levels of Cyclin D1 and Bcl‐2, which are required for cell cycle progression and cell proliferation.24, 30
Epidemiological data strongly suggest that the heavy drinking increases the risk for pancreatic cancer.6, 10, 31, 32 High alcohol intake was associated with a higher risk of pancreatitis.33–35 Diabetic patients and heavy smokers who consume excessive alcohol demonstrate a high risk of developing pancreatic cancer. In compared to pancreatitis, the role of alcohol consumption remains less clear in pancreatic cancer. A low to moderate alcohol consumption does not appear to be associated with pancreatic cancer risk, and only chronic heavy drinking increases the risk of pancreatic cancer. Several factors have been demonstrated to contribute towards the development of alcohol‐associated cancer.10, 36–39 Evidence suggests that the effect of alcohol is modulated by polymorphisms in genes encoding enzymes for ethanol metabolism (eg alcohol dehydrogenases, aldehyde dehydrogenases and cytochrome P450 2E1), folate metabolism and DNA repair. Several events including a genotoxic effect of acetaldehyde increased oestrogen concentration, a role as the solvent for tobacco carcinogens, production of ROS and nitrogen species, and changes in folate metabolism have been implicated. Daily consumption of more than 80 g alcohol (more than five to six drinks) with smoking increases the risk of developing cancers by a factor of 50 or more.40, 41 In support of our study, it was demonstrated that chronic alcohol consumption promotes intestinal tumorigenesis and tumour invasion in genetically susceptible mice.15 During ethanol metabolism, ethanol is oxidized to acetaldehyde by ADH or CYP2E1.42, 43 If the patients consume alcohol, acetaldehyde concentrations in the stomach increase 6.5‐fold.44 In addition to the acetaldehyde generated by cellular enzymes or gastrointestinal bacteria, considerable amounts of acetaldehyde are present in certain alcoholic beverages and cigarette smoke.44 Based on these findings it appears that ethanol and its metabolites are the potential human carcinogens.
SATB2 may act as a master regulator of pluripotency and self‐renewal because SATB2 binding sites are present in the promoter regions of KLF4, Oct4, cMyc and Sox2.45–48 Similar to pancreatic CSCs, we have found that SATB2 is highly expressed in colorectal and breast CSCs.24, 30 Interestingly, the expression of SATB2 was absent or very low in HPNE cells, mammary epithelial cells and colorectal epithelial cells.20, 24, 30 Furthermore, overexpression of SATB2 in normal epithelial cells resulted in malignant transformation, suggesting an oncogenic role of SATB2 in various cancers. In the present study, ethanol‐transformed cells express high levels of SATB2 which was associated with generation of ROS. At present the basis of SATB2 induction by chronic ethanol exposure to HPNE cells is not clear. It is possible that chronic ethanol exposure of HPNE cells causes ROS production (an inflammatory environment) which in turn induces SATB2 expression and malignant transformation. Alternatively, other oncogenes are simultaneously amplified to sustain‐transformed phenotypes of CSCs, and high level of ROS is an outcome of transformation.
In conclusion, our data demonstrate that chronic ethanol exposure of HPNE cells induced malignant transformation, generated ROS, and these transformed cells gained the phenotype of CSCs by expressing SATB2, stem cell markers, and pluripotency maintaining factors. These findings are very significant for human health because alcohol can initiate or promote signalling events, either alone or in combination with other unknown factors, which could result in the development of pancreatic cancer. Further studies are underway in our laboratory to fully understand the pathological significance of alcohol toxicity in mouse models of pancreatic cancer. Overall, precautions should be taken when alcohol is consumed continuously for a longer period.
ACKNOWLEDGEMENTS
The authors acknowledge all the laboratory members for critical reading, assistance and suggestions during manuscript preparation. This work was supported by a grant (R01AA024698) from the National Institutes of Health, USA.
CONFLICTS OF INTEREST
All the authors have declared that no conflict of interest exist.
AUTHOR CONTRIBUTIONS
WY and YM performed the experiments, analysed the data and wrote the manuscript. SS, and RKS designed the study and contributed reagents. All authors read and approved the manuscript.
REFERENCES
- 1Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018; 68: 7‐ 30.Wiley Online Library PubMed Web of Science®Google Scholar
- 2Warshaw AL, Fernandez‐del Castillo C. Pancreatic carcinoma. N Engl J Med. 1992; 326: 455‐ 465.Crossref PubMed Web of Science®Google Scholar
- 3Magee CJ, Ghaneh P, Neoptolemos JP. Surgical and medical therapy for pancreatic carcinoma. Best Pract Res Clin Gastroenterol. 2002; 16: 435‐ 455.Crossref PubMed Web of Science®Google Scholar
- 4Singh D, Upadhyay G, Srivastava RK, Shankar S. Recent advances in pancreatic cancer: biology, treatment, and prevention. Biochim Biophys Acta. 2015; 1856: 13‐ 27.Crossref CAS PubMed Web of Science®Google Scholar
- 5Bardeesy N, DePinho RA. Pancreatic cancer biology and genetics. Nat Rev Cancer. 2002; 2: 897‐ 909.Crossref CAS PubMed Web of Science®Google Scholar
- 6Li D, Jiao L. Molecular epidemiology of pancreatic cancer. Int J Gastrointest Cancer. 2003; 33: 3‐ 14.Crossref Web of Science®Google Scholar
- 7Raimondi S, Maisonneuve P, Lowenfels AB. Epidemiology of pancreatic cancer: an overview. Nat Rev Gastroenterol Hepatol. 2009; 6: 699‐ 708.Crossref PubMed Web of Science®Google Scholar
- 8Whitcomb DC, Pogue‐Geile K. Pancreatitis as a risk for pancreatic cancer. Gastroenterol Clin North Am. 2002; 31: 663‐ 678.Crossref PubMed Web of Science®Google Scholar
- 9Yadav D, Lowenfels AB. The epidemiology of pancreatitis and pancreatic cancer. Gastroenterology. 2013; 144: 1252‐ 1261.Crossref PubMed Web of Science®Google Scholar
- 10Herreros‐Villanueva M, Hijona E, Banales JM, Cosme A, Bujanda L. Alcohol consumption on pancreatic diseases. World J Gastroenterol. 2013; 19: 638‐ 647.Crossref PubMed Web of Science®Google Scholar
- 11Ahlgren JD. Epidemiology and risk factors in pancreatic cancer. Semin Oncol. 1996; 23: 241‐ 250.PubMed Web of Science®Google Scholar
- 12Duell EJ. Epidemiology and potential mechanisms of tobacco smoking and heavy alcohol consumption in pancreatic cancer. Mol Carcinog. 2012; 51: 40‐ 52.Wiley Online Library CAS PubMed Web of Science®Google Scholar
- 13Gupta S, Wang F, Holly EA, Bracci PM. Risk of pancreatic cancer by alcohol dose, duration, and pattern of consumption, including binge drinking: a population‐based study. Cancer Causes Control. 2010; 21: 1047‐ 1059.Crossref PubMed Web of Science®Google Scholar
- 14Longnecker MP, Enger SM. Epidemiologic data on alcoholic beverage consumption and risk of cancer. Clin Chim Acta. 1996; 246: 121‐ 141.Crossref CAS PubMed Web of Science®Google Scholar
- 15Wimberly AL, Forsyth CB, Khan MW, Pemberton A, Khazaie K, Keshavarzian A. Ethanol‐induced mast cell‐mediated inflammation leads to increased susceptibility of intestinal tumorigenesis in the APC Delta468 min mouse model of colon cancer. Alcohol Clin Exp Res. 2013; 37(Suppl 1): E199‐ E208.Wiley Online Library CAS PubMed Web of Science®Google Scholar
- 16Dobreva G, Dambacher J, Grosschedl R. SUMO modification of a novel MAR‐binding protein, SATB2, modulates immunoglobulin mu gene expression. Genes Dev. 2003; 17: 3048‐ 3061.Crossref CAS PubMed Web of Science®Google Scholar
- 17Cai S, Han HJ, Kohwi‐Shigematsu T. Tissue‐specific nuclear architecture and gene expression regulated by SATB1. Nat Genet. 2003; 34: 42‐ 51.Crossref CAS PubMed Web of Science®Google Scholar
- 18Cai S, Lee CC, Kohwi‐Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet. 2006; 38: 1278‐ 1288.Crossref CAS PubMed Web of Science®Google Scholar
- 19Dobreva G, Chahrour M, Dautzenberg M, et al. SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell. 2006; 125: 971‐ 986.Crossref CAS PubMed Web of Science®Google Scholar
- 20Yu W, Ma Y, Shankar S, Srivastava RK. Role of SATB2 in human pancreatic cancer: implications in transformation and a promising biomarker. Oncotarget. 2016; 7: 57783‐ 57797.Crossref PubMed Google Scholar
- 21Nalls D, Tang SN, Rodova M, Srivastava RK, Shankar S. Targeting epigenetic regulation of miR‐34a for treatment of pancreatic cancer by inhibition of pancreatic cancer stem cells. PLoS ONE. 2011; 6: e24099.Crossref CAS PubMed Web of Science®Google Scholar
- 22Fu J, Rodova M, Roy SK, et al. GANT‐61 inhibits pancreatic cancer stem cell growth in vitro and in NOD/SCID/IL2R gamma null mice xenograft. Cancer Lett. 2013; 330: 22‐ 32.Crossref CAS PubMed Web of Science®Google Scholar
- 23Asara Y, Marchal JA, Carrasco E, et al. Cadmium modifies the cell cycle and apoptotic profiles of human breast cancer cells treated with 5‐Fluorouracil. Int J Mol Sci. 2013; 14: 16600‐ 16616.Crossref PubMed Web of Science®Google Scholar
- 24Yu W, Ma Y, Shankar S, Srivastava RK. SATB2/beta‐catenin/TCF‐LEF pathway induces cellular transformation by generating cancer stem cells in colorectal cancer. Sci Rep. 2017; 7: 10939.Crossref PubMed Web of Science®Google Scholar
- 25Ma Y, Yu W, Shrivastava A, et al. Sanguinarine inhibits pancreatic cancer stem cell characteristics by inducing oxidative stress and suppressing sonic hedgehog‐Gli‐Nanog pathway. Carcinogenesis. 2017; 38: 1047‐ 1056.Crossref PubMed Web of Science®Google Scholar
- 26Shankar S, Singh G, Srivastava RK. Chemoprevention by resveratrol: molecular mechanisms and therapeutic potential. Front Biosci. 2007; 12: 4839‐ 4854.Crossref CAS PubMed Web of Science®Google Scholar
- 27Singh BN, Fu J, Srivastava RK, Shankar S. Hedgehog signaling antagonist GDC‐0449 (Vismodegib) inhibits pancreatic cancer stem cell characteristics: molecular mechanisms. PLoS ONE. 2011; 6: e27306.Crossref CAS PubMed Web of Science®Google Scholar
- 28Tang SN, Fu J, Nall D, Rodova M, Shankar S, Srivastava RK. Inhibition of sonic hedgehog pathway and pluripotency maintaining factors regulate human pancreatic cancer stem cell characteristics. Int J Cancer. 2012; 131: 30‐ 40.Wiley Online Library CAS PubMed Web of Science®Google Scholar
- 29Rosenfeld JA, Ballif BC, Lucas A, et al. Small deletions of SATB2 cause some of the clinical features of the 2q33.1 microdeletion syndrome. PLoS ONE. 2009; 4: e6568.Crossref CAS PubMed Web of Science®Google Scholar
- 30Yu W, Ma Y, Ochoa AC, Shankar S, Srivastava RK. Cellular transformation of human mammary epithelial cells by SATB2. Stem Cell Res. 2017; 19: 139‐ 147.Crossref CAS PubMed Web of Science®Google Scholar
- 31Longnecker MP. Alcohol consumption and risk of cancer in humans: an overview. Alcohol. 1995; 12: 87‐ 96.Crossref PubMed Web of Science®Google Scholar
- 32Michaud DS, Vrieling A, Jiao L, et al. Alcohol intake and pancreatic cancer: a pooled analysis from the pancreatic cancer cohort consortium (PanScan). Cancer Causes Control. 2010; 21: 1213‐ 1225.Crossref PubMed Web of Science®Google Scholar
- 33DiMagno MJ, DiMagno EP. Chronic pancreatitis. Curr Opin Gastroenterol. 2010; 26: 490‐ 498.Crossref PubMed Web of Science®Google Scholar
- 34Go VL, Gukovskaya A, Pandol SJ. Alcohol and pancreatic cancer. Alcohol. 2005; 35: 205‐ 211.Crossref CAS PubMed Web of Science®Google Scholar
- 35Nitsche C, Simon P, Weiss FU, et al. Environmental risk factors for chronic pancreatitis and pancreatic cancer. Dig Dis. 2011; 29: 235‐ 242.Crossref PubMed Web of Science®Google Scholar
- 36Breeden JH. Alcohol, alcoholism, and cancer. Med Clin North Am. 1984; 68: 163‐ 177.Crossref CAS PubMed Web of Science®Google Scholar
- 37Kocher HM, Alrawashdeh W. Pancreatic cancer. BMJ Clin Evid. 2010; 2010. pii: 0409.PubMed Google Scholar
- 38Ringborg U. Alcohol and risk of cancer. Alcohol Clin Exp Res. 1998; 22: 323S‐ 328S.Wiley Online Library CAS PubMed Web of Science®Google Scholar
- 39Stermer E. Alcohol consumption and the gastrointestinal tract. Isr Med Assoc J. 2002; 4: 200‐ 202.PubMed Web of Science®Google Scholar
- 40Poschl G, Seitz HK. Alcohol and cancer. Alcohol Alcohol. 2004; 39: 155‐ 165.Crossref CAS PubMed Web of Science®Google Scholar
- 41Poschl G, Stickel F, Wang XD, Seitz HK. Alcohol and cancer: genetic and nutritional aspects. Proc Nutr Soc. 2004; 63: 65‐ 71.Crossref CAS PubMed Web of Science®Google Scholar
- 42Leung TM, Nieto N. CYP2E1 and oxidant stress in alcoholic and non‐alcoholic fatty liver disease. J Hepatol. 2013; 58: 395‐ 398.Crossref CAS PubMed Web of Science®Google Scholar
- 43Elamin EE, Masclee AA, Dekker J, Jonkers DM. Ethanol metabolism and its effects on the intestinal epithelial barrier. Nutr Rev. 2013; 71: 483‐ 499.Wiley Online Library PubMed Web of Science®Google Scholar
- 44Kurkivuori J, Salaspuro V, Kaihovaara P, et al. Acetaldehyde production from ethanol by oral streptococci. Oral Oncol. 2007; 43: 181‐ 186.Crossref CAS PubMed Web of Science®Google Scholar
- 45Asanoma K, Kubota K, Chakraborty D, et al. SATB homeobox proteins regulate trophoblast stem cell renewal and differentiation. J Biol Chem. 2012; 287: 2257‐ 2268.Crossref CAS PubMed Web of Science®Google Scholar
- 46Conner JR, Hornick JL. SATB2 is a novel marker of osteoblastic differentiation in bone and soft tissue tumours. Histopathology. 2013; 63: 36‐ 49.Wiley Online Library PubMed Web of Science®Google Scholar
- 47Gyorgy AB, Szemes M, de Juan Romero C, Tarabykin V, Agoston DV. SATB2 interacts with chromatin‐remodeling molecules in differentiating cortical neurons. Eur J Neurosci. 2008; 27: 865‐ 873.Wiley Online Library PubMed Web of Science®Google Scholar
- 48Szemes M, Gyorgy A, Paweletz C, Dobi A, Agoston DV. Isolation and characterization of SATB2, a novel AT‐rich DNA binding protein expressed in development‐ and cell‐specific manner in the rat brain. Neurochem Res. 2006; 31: 237‐ 246.Crossref CAS PubMed Web of Science®Google Scholar
- 49Shankar S, Nall D, Tang SN, et al. Resveratrol inhibits pancreatic cancer stem cell characteristics in human and KrasG12D transgenic mice by inhibiting pluripotency maintaining factors and epithelial‐mesenchymal transition. PLoS ONE. 2011; 6: e16530.Crossref CAS PubMed Web of Science®Google Scholar
- 50Britanova O, Akopov S, Lukyanov S, Gruss P, Tarabykin V. Novel transcription factor Satb2 interacts with matrix attachment region DNA elements in a tissue‐specific manner and demonstrates cell‐type‐dependent expression in the developing mouse CNS. Eur J Neurosci. 2005; 21: 658‐ 668.Wiley Online Library CAS PubMed Web of Science®Google Scholar
Citing Literature
Number of times cited according to CrossRef: 4
- Ilaria Chiodi, Chiara Mondello, Life style factors, tumor cell plasticity and cancer stem cells, Mutation Research/Reviews in Mutation Research, 10.1016/j.mrrev.2020.108308, (108308), (2020).Crossref
- Wei Yu, Sanjit K. Roy, Yiming Ma, Thomas A. LaVeist, Sharmila Shankar, Rakesh K. Srivastava, Higher expression of SATB2 in hepatocellular carcinoma of African Americans determines more aggressive phenotypes than those of Caucasian Americans, Journal of Cellular and Molecular Medicine, 10.1111/jcmm.14652, 23, 12, (7999-8009), (2019).Wiley Online Library
- Arkadiusz Gzil, Izabela Zarębska, Wiktor Bursiewicz, Paulina Antosik, Dariusz Grzanka, Łukasz Szylberg, Markers of pancreatic cancer stem cells and their clinical and therapeutic implications, Molecular Biology Reports, 10.1007/s11033-019-05058-1, (2019).Crossref
- Suthipong Chujan, Tawit Suriyo, Jutamaad Satayavivad, Integrative In Silico and In Vitro Transcriptomics Analysis Revealed Gene Expression Changes and Oncogenic Features of Normal Cholangiocytes after Chronic Alcohol Exposure, International Journal of Molecular Sciences, 10.3390/ijms20235987, 20, 23, (5987), (2019).Crossref